The Johnson lab is primarily interested in mechanisms of small GTPase function in disease, with a particular emphasis on cancer. However, we invite collaboration and communication with any lab inside and outside the University of South Carolina community interested in protein chemistry and structural biology.
Project 1: GTPase autophosphorylation
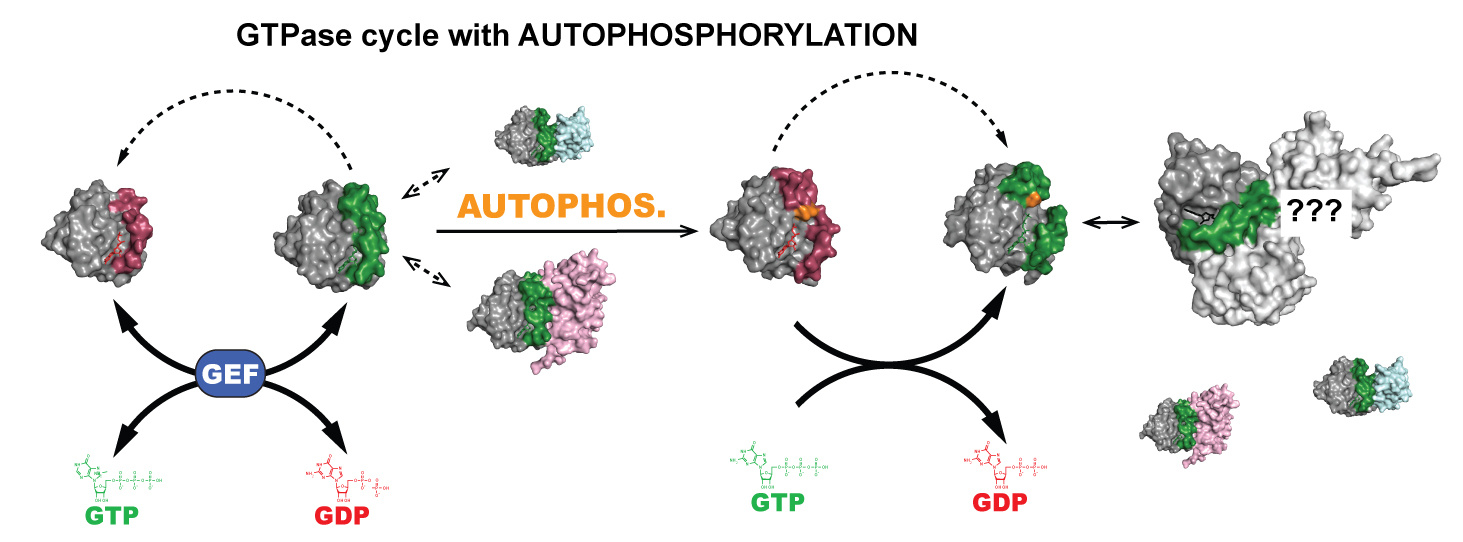
It has been known for ~50 years that active site mutations in GTPases can switch these
enzymes from favoring GTP hydrolysis to phosphoryl-transfer. However, the mechanism
and biological function of this mutation-induced change in enzyme function is poorly
understood. Surprisingly, several other small GTPases show potential for autophosphorylation,
suggesting that this reaction has conserved yet unknown cellular functions. The goal of this project is to 1) understand the molecular mechanism of autophosphorylation
and how it competes with GTP hydrolysis, 2) determine the extent of autophosphorylation
as a conserved evolutionary function of wildtype small GTPases and its biological
function(s), 3) determine how this post-translational modification influences GTPase
interactions with other proteins, 4) specify the molecular interactions that allow
auto-phosphorylated small GTPases to transform cells and participate in oncogenesis,
and 5) explore autophosphorylation as a functional tool to study other members of
the small GTPase superfamily.
Project 2: Mechanisms of GTPase nucleotide exchange
Activation of small GTPases require the exchange of GDP for GTP. For KRAS, HRAS, and
NRAS (collectively RAS), enhancement of nucleotide exchange by guanine nucleotide
exchange factors (GEFs) is well described structurally. The same is not true for the
process of intrinsic nucleotide exchange, whereby exchange of GDP for GTP occurs without the aid of a
partner GEF to bind the active site of RAS to catalyze nucleotide release. The reason
why the processes of intrinsic nucleotide exchange has been largely ignored is because
the exchange process occurs very slowly. However, many oncogenic and germline mutations of RAS proteins promote nucleotide exchange
as a primary mechanism of activation. Therefore, a central question is whether 1) intrinsic nucleotide exchange in wildtype
RAS and its mutants occur through a common change in protein conformation to allow
release and binding of different nucleotides, or through a spectrum of conformational
transitions? Further, 2) are these conformational transitions unique compared to GEF-mediated
exchange, or do GEF and intrinsic nucleotide exchange mechanisms occur via a common
conformational transition? While answering these questions are important for a better
understanding of mutant RAS function, they’re also critical for the 3) design of chemical
probes to investigate specific nucleotide-exchange promoting mutants of RAS and their
role in pathogenesis.
Project 3: Functional characterization of GTPase-RASSF interactions
The Ras-association domain family (RASSF) are a large group of putative tumor suppressors
and scaffold proteins that are potentially regulated by small GTPases. All members
of the RASSF family contain RalGDS/AF6 Ras association (RA) domains capable of binding
the G-domain of small GTPases. A prominent example is the KRAS-RASSF5 complex, which
has been shown by several groups to suppress cell growth in culture, and which appears
to do so through the generation of several different ternary complexes. The molecular nature of these complexes, and their relevance for disease progression in vivo, remains to be elucidated. For instance, greater than half of all RASSF proteins form constitutive complexes
with the evolutionarily conserved Hippo kinases MST1/2 which are critical for organ
homeostasis, and this is true for RASSF5 as well, but how KRAS participates in MST1/2 function via RASSF5 is not understood. Therefore, the specific goals of this project are to 1) decipher the molecular interactions
between KRAS, RASSF5, and MST1/2 proteins, 2) decode KRAS/RASSF5 interaction partners
outside of MST1/2, 3) test the tumor suppressor functions of KRAS/RASSF in in vivo models of colon tumorigenesis. Further, besides KRAS and a few outstanding examples
(e.g. GEM, RAP1), most other RASSF proteins lack an identified cognate GTPase. Thus,
we will expand the knowledge of RASSF function by 4) identifying, validating, and
characterizing at an atomic level other RASSF-GTPase pairs. In doing so, we will elucidate
the basic functionality of RASSF-GTPase complexes at the atomic, molecular, and biological
levels.
Project 4: Intra-cooperation between oncogenes and tumor suppressors of the MAPK signaling pathway

Oncogenic mutations of KRAS contribute to colorectal cancer progression through activation of the RAF/MEK/ERK signaling pathway. Traditionally, co-occurring mutations in KRAS and other genes that regulate the MAPK signaling pathway have been viewed as classic examples of negative epistasis, whereby cancer cells select against the presence of multiple MAPK pathway activating mutations due to cellular toxicity. However, oncogenic mutations of KRAS are heterogenous, and KRAS allele heterogeneity in the context of genetic epistasis is understudied. In fact, some oncogenic KRAS alleles appear to favor co-mutations in genes of the MAPK-signaling pathway normally viewed as mutually exclusive. In this project, we seek to move the field beyond phenomenological descriptions of epistasis to 1) understanding the protein-protein interactions that define these relationships, 2) modelling of these interactions in vivo to understand their contribution to colorectal cancer, and lastly 3) testing their contributions to both primary and acquired drug resistance.